Two years into a $49.5 million cancer-mapping project, researchers are opening the door to new kinds of tests that could alert doctors to multiple kinds of cancer when they’re most treatable.
(text and background only visible when logged in)
When a Georgia Tech-led project received a contract award from the Advanced Research Projects Agency for Health (ARPA-H), it was for a bold idea with aggressive metrics. And it wasn’t guaranteed money. The team, led by biomedical engineer Gabe Kwong, had to deliver on its vision. Doing so could transform cancer screening and care, leading to one-size-fits-all tests that detect multiple cancers before they’re visible on CT or PET scans.
It’s a big goal, but that’s the point of ARPA-H. The agency funds staggeringly difficult healthcare innovation ideas that require major investment to succeed.
Two years into the $49.5 million project, Kwong and the team from Georgia Tech, Columbia University, and Mount Sinai Health System has crossed a critical threshold.
They’ve built the first tool able to measure enzyme activity around cancer tumors and healthy cells. And they’ve deployed it to understand the unique signatures for tumors from 14 different kinds of cancer.
That data is powering the first version of a cancer “atlas.” Like a geographical atlas, it will offer directions to each kind of tumor, allowing scientists to design sensors that follow the map and detect cancer tumors when they’re still small.
“If I want to deliver a sensor to a particular region inside the body, right now, there's no way of directing it. We give it systemically, and it basically infuses all tissues all the time,” said Kwong, Robert A. Milton Professor in the Wallace H. Coulter Department of Biomedical Engineering. “What's powerful is that we’re now defining tissue sites with a specific molecular ‘barcode.’ Then if a sensor is given systemically, it should only turn on when the barcode matches the local tissue.”
Because the ideas are big and failure can happen, an ARPA-H contract can end at any time. Researchers must keep meeting their targets so ultimately, their work can be pitched to investors, commercialized, and become a solution to help patients.
Meeting these first milestones has earned the team approval to continue. It’s also unlocked continued funding and is one of the first ARPA-H projects to transition into the second phase of work.
ARPA-H Program Manager Ross Uhrich, DMD, MBA, who oversees the Cancer and Organ Degradome Atlas (CODA) project said: “In the ARPA-H model, selection for continued funding is by no means a guarantee. Progression to Phase II and distribution of follow-on capital are direct reflections of a project team’s successful performance across rigorous technical, commercial, and regulatory metrics.
Two Big Wins

Biomedical engineer Gabe Kwong says his ARPA-H project has accelerated a decade's worth of science into a handful of years, bringing a future of multi-cancer screening tools closer than ever.
About 40% of people in the U.S. will develop cancer in their lifetimes, according to the American Cancer Society. Often, it’s detected in a later stage, when treatment can be expensive and difficult.
Catching tumors sooner would keep people healthier, improve quality of life, and reduce the costs of healthcare.
Yet effective cancer screenings are few and limited, and they can only find one kind of disease. For example, colonoscopies check for colorectal cancer, mammograms for breast cancer, and prostate-specific antigen (PSA) tests for prostate cancer.
If a single test could find multiple diseases, doctors could catch cancer in its early stages, when it’s small and most effectively treated.
But so far, the hunt for signs of growing tumors has depended on finding tell-tale natural biomarkers, such as DNA or proteins. These are often found in vanishingly small quantities and difficult to detect, Kwong said. They’re also produced by normal biological activity, resulting in false alarms.
With CODA, new sensors could actively search and release easily detectable synthetic molecules when they encountered a cancer’s specific enzyme activity.
No map of cancer activity like this exists.
“The fundamental premise is that the address for breast cancer is different from the address for lung cancer, which is different from the address for healthy lung tissue,” Kwong said.
“The first-generation data will include several models of each cancer type, which shows that our idea works. The expectation is that over time, the atlas will be a living document; as we get more information, it will grow.”
Kwong and his team first had to prove it was possible to catalog the enzyme activity around tumors and healthy tissue.
They’ve done that. And they’re now profiling — creating the barcodes — for 14 types of cancer.
“If that tool did not work, ARPA-H would not be continuing to fund our work,” Kwong said. “We proved we could assemble the atlas because we proved our measurement tool can do exactly what we thought it could do.”
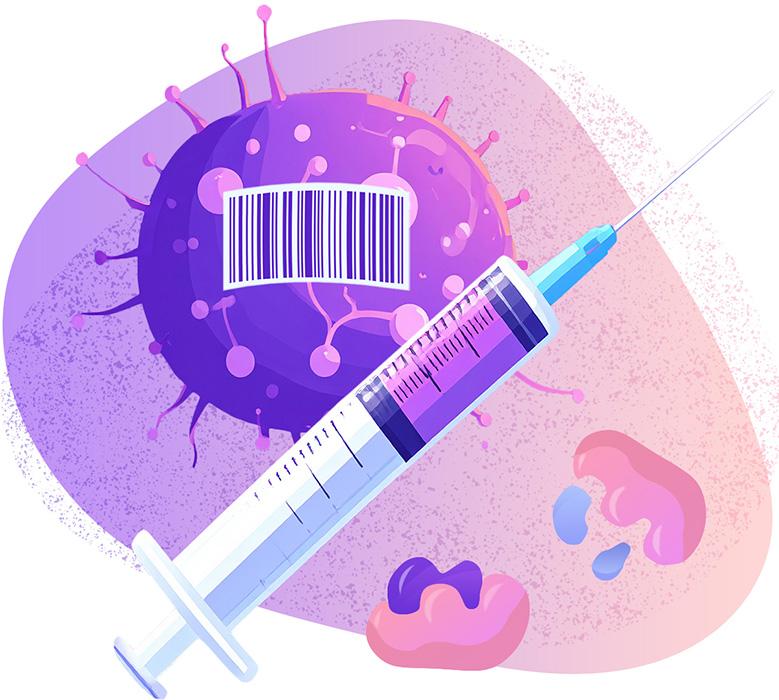
With a Map, New Sensors Take Shape
In the second phase of their work, the Georgia Tech-led team will finalize the atlas and put their initial sensor designs through rigorous lab tests. So far, three different sensors are taking shape, all using molecular “logic” to sense and respond to tumor cells.
Just like computers require a username, password, and multifactor authentication to access systems, these sensors would require multiple kinds of enzyme activity at the same location in the body to send up an alert.
“Their preliminary studies in preclinical models are very promising. The team’s prototype sensors already outperform market-comparable biomarkers in detecting small tumors,” Uhrich said. “We look forward to supporting their Phase II activities, which will bring them one step closer to their goal of creating a first-of-its-kind atlas of common cancers and using that atlas to generate highly specific, lab-based early cancer screening tests that can revolutionize the market.”
Kwong is working with Georgia Tech researchers John Blazeck in the School of Chemical and Biomolecular Engineering and Peng Qiu in Coulter BME on the project. Their key collaborators include Columbia biomedical engineer Tal Danino and Min Xue at Mount Sinai. Eventually, the atlas will be available to other researchers with ideas for new cancer screening tools, too.
If it all sounds a little like science fiction, Kwong would suggest that’s the point. ARPA-H’s model has accelerated a decade’s worth of scientific work in multi-cancer screening into two years.
“It's scope and scale,” he said. “If I did a typical research approach, I’m not sure I would ever get there in 20-30 years. This is shrinking all that and doing the work in three to five years.”
(text and background only visible when logged in)
Related Content

$50M Cancer Moonshot Grant Will Build an Atlas for Earlier Cancer Detection
Biomedical engineer Gabe Kwong will map cancer cell biomarkers, then engineer new sensors to hunt for multiple kinds of cancer.
Biomedical engineer Gabe Kwong will map cancer cell biomarkers, then engineer new sensors to hunt for multiple kinds of cancer.

BME Researchers Lead $24M Project Using mRNA to ‘Turn On’ Helpful Immune Responses
Philip Santangelo wants to build a toolbox of mRNA drugs to activate or shut off specific genes to help the immune system fight cancer and other disorders.
Philip Santangelo wants to build a toolbox of mRNA drugs to activate or shut off specific genes to help the immune system fight cancer and other disorders.

Painting a Target on Cancer to Make Therapy More Effective
A BME team is putting a synthetic flag on tumors, then engineering a patient’s immune cells to find and eliminate cancer.
A BME team is putting a synthetic flag on tumors, then engineering a patient’s immune cells to find and eliminate cancer.